Functional Genetics of Movement Disorders

Team: Ana Westenberger, PhD (group leader); Aloysius Domingo, MD (visiting scientist, PhD student); Karen Grütz, née Freimann, PhD (Postdoc); Heike Pawlack (research technologist); Thomas Schmidt (MD student); and Christoph Max (MD student)
The research team “Functional genetics of movement disorders” focuses on the identification and functional characterization of the structural and epigenetic DNA changes that lead to the development of various movement disorders. To achieve these goals, we combine linkage analysis with Sanger and next-generation sequencing and real-time quantitative PCR. The identification of DNA changes is followed by a series of molecular biology, cellular biology, and protein biochemistry assays. By this, we examine the consequences of the detected DNA changes on transcriptional regulation of gene expression and on protein translation levels, subcellular localization, stability, and functional interactions of the affected proteins. Finally, we aim to correlate our findings with the observed clinical phenotypes and hypothesize mechanism(s) of disease.
Importantly, together with other members of the ProtectMove consortium, we focus on identifying genetic and epigenetic factors that modify the penetrance and expressivity of various neurogenetic disorders.
Our current projects investigate the genetic basis of X-linked dystonia-parkinsonism (XDP), primary familial brain calcification (PFBC), ADCY5-related movement disorder, and other neurogenetic conditions.
In addition, we are intensively involved in development, data extraction, and curation of the MDSGene database.
We value and acknowledge scientific exchange and input from our international collaborators Prof. Dr. Henry L. Paulson (University of Michigan, USA), Prof. Dr. Lillian V. Lee and Prof. Dr. Raymond Rosales (XDP study group, Philippines), and Prof. Dr. Vladimir S. Kostic (University of Belgrade, Serbia).
Our projects are funded by the German Research Foundation, Fritz-Thyssen Foundation, and intramural grant support.
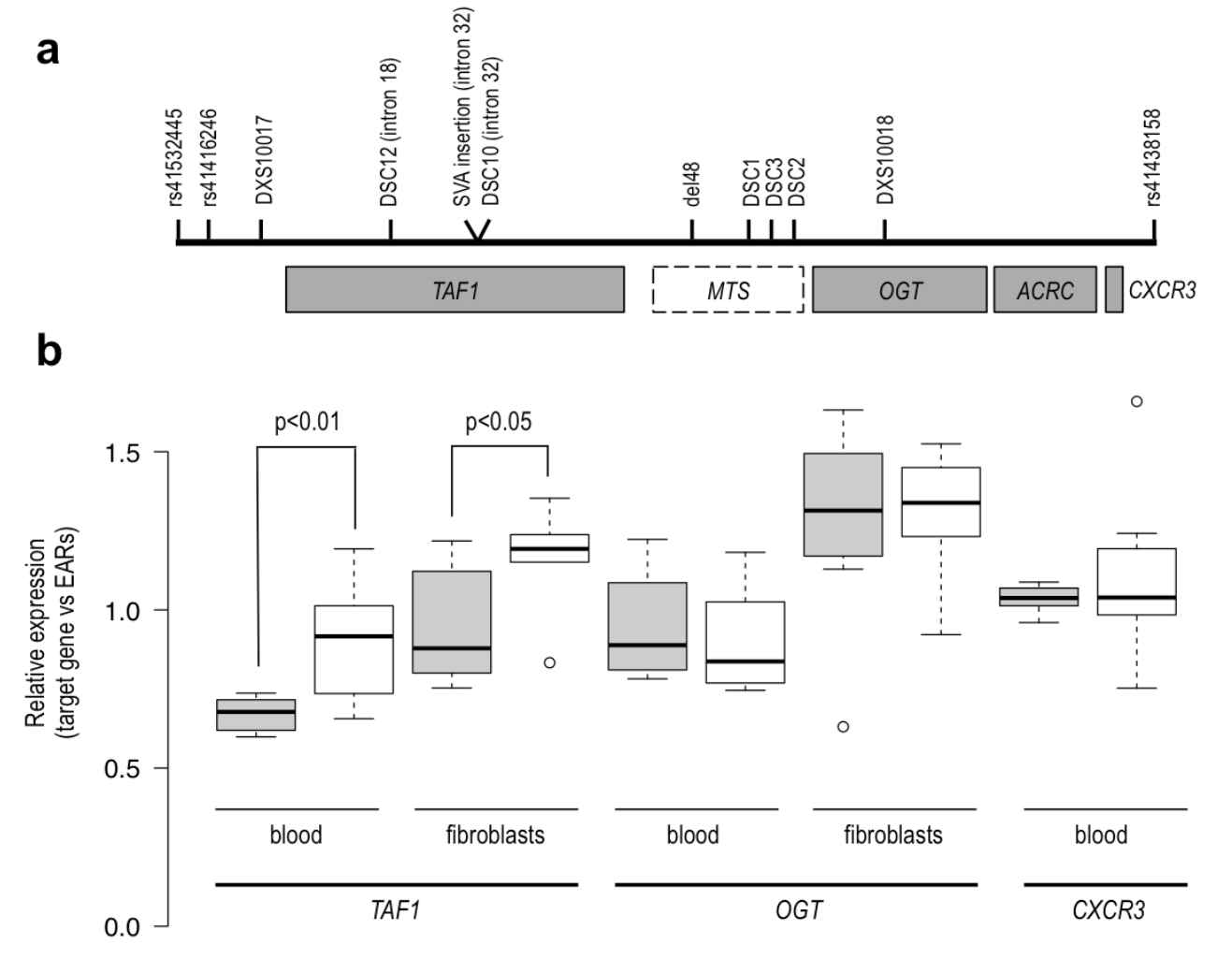

A. Linked region in Xq13.1 showing XDP-associated genetic changes (“XDP haplotype”: DSC12, SVA retrotransposon insertion, DSC10, del48, DSC1, DSC2, DSC3), located either in introns of TAF1 or within an untranslated region (“Multiple Transcript System”, MTS) distal to the gene.
B. qPCR analysis showing significant downregulation of TAF1 in the XDP group (shaded boxes) in both blood and fibroblast-derived RNA, and no difference in two other genes in the linked region. TAF1 and OGT are both expressed in blood and fibroblasts, while CXCR3 is expressed only in blood. Boxplot whiskers extend to data points that are within 1.5x the interquartile ranges. EARs – Expressed Alu Repeats. (Domingo et al., Cell Mol Life Sci. 2016).
- Translational Neurogenetics
- Genetics of Rare Diseases
- Functional Genetics of Movement Disorders
- Integrative Omics in Parkinson Disease
- Neuropsychiatric Epidemiology
- Clinical Neurogenetics and Neuroimaging
- Mitochondrial function in movement disorders
- Institut für Systemische Motorikforschung
- Applied Stem Cell Biology